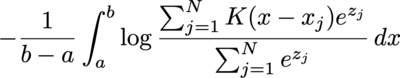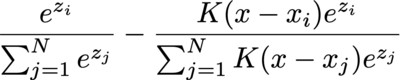Property of gradient of cross-entropy loss with kernel density estimation
← Thread:Talk:BeepBoop/Understanding BeepBoop/Property of gradient of cross-entropy loss with kernel density estimation
I'm quite curious about the behavior of cross entropy loss between a uniform distribution and kernel density estimation with softmax weight:
where a and b is the lower and upper bound of the target uniform distribution, K is the kernel function (assume normalized), x_j is the angle of the data point, and z_j the weight before softmax.
The integral is often calculated by numerical methods, such as binning, so let's consider the gradient of the i-th data point's weight before softmax, and consider only one of the bins (with angle x) and ignore the values multiplied before integral:
It degenerates to ordinary cross entropy loss with softmax when K is either 1 or 0 (and 1 iif the "label" matches): S_i - 1 when label matches or S_i when label mismatches.
But things start to get interesting when K is different.
Interesting observations! The scale invariance of K actually seems like a good property to me. It means that K doesn't really need to be normalized, or more precisely that multiplying K by a constant multiplies the gradient by that constant, which seems like the behavior you'd want. Most loss functions (e.g., mean squared error) learn more for far data points than close ones. That might be good for surfing, but I could imagine that you may want the opposite for targeting where you "give up" on hard data points and focus on the ones that you might score a hit on. So perhaps BeepBoop's loss is a middle ground that works decently well for both.
One more finding. Actually you don't need to take integral or use bins at all, you can compute the loss from each data point separately and take the sum of the loss. Although the value in loss isn't equal, the gradients are exactly the same. This yields one more insight: the absolute predicted value isn't important at all, all that matters is how relatively they are close to the target distribution, compared to each other. As a result, the cluster used for one prediction isn't necessarily in the same batch, they can be shuffled entirely, yet doesn't affect the result (theoretically).
Oops the calc is wrong.